You are here
Pathogen genetic diversity and evolution
Within a line of investigation on population and evolutionary genetics of both pathogens (H. vastatrix and C. kahawae), we have been using genetic markers and genomic approaches to: i) investigate patterns of pathogen population diversity, differentiation and distribution to understand epidemiological dynamics and their evolutionary history; ii) identify candidate genes and genomic mechanisms responsible for host adaptation and virulence evolution; iii) identify molecular markers associated with specific pathotypes.
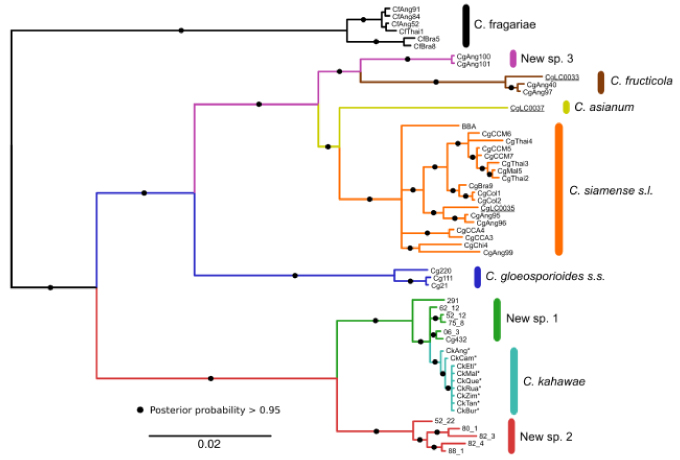
Our previous works led to the development of new DNA markers for the Colletotrichum gloesporioides species complex, which proved fundamental to distinguish closely related Colletotrichum species and unequivocally identify C. kahawae. A subsequent population study revealed a geographically related genetic structuring in C. kahawae and its probable geographical origin, and enabled the proposal of a new hypothesis of speciation through host-jump, which contradicted the long standing previous hypotheses.
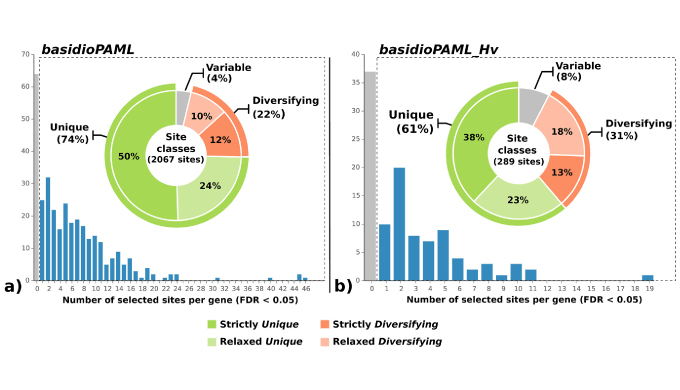
A genomic study to investigate the evolutionary origin of rust fungi (to which H. vastatrix belongs) was conducted through a genome-wide scan for positive selection and genes undergoing accelerated evolution, integrated into a phylogenomics framework. From this approach, the role of positive selection at the origin and evolution of rust fungi was evidenced, revealing targeted candidate genes that may have been involved in evolutionary change, within which a significant predominance of genes involved in amino acid transport and metabolism was found.
[Silva et al. (2015) PLoS ONE 10(12):e0143959 (doi:10.1371/journal.pone.0143959)]
Currently, we are undertaking population genomic studies for a large range of both H. vastatrix and C. kahawae isolates.
Selected References:
- Silva et al. (2018). Molecular Plant Pathology (Accepted manuscript online). (DOI: 10.1111/mpp.12657).
- Batista et al. (2017). Frontiers in Plant Science 7: 2051 (DOI: 10.3389/fpls.2016.02051).
- Talhinhas et al. (2017). Molecular Plant Pathology 18 (8): 1039–1051 (DOI: 10.1111/mpp.12512)
- Vieira et al. (2016) PLoS ONE 11(3): e0150651 (doi: 10.1371/journal.pone.0150651)
- Silva et al. (2015) PLoS ONE 10(12):e0143959 (doi:10.1371/journal.pone.0143959)
- Batista et al. (2015) In: Proceedings of the 25th International Conference on Coffee Science, Association for Science and Information on Coffee (ASIC) [Eds.], Armenia, Colombia, pp: 42-46 pb 220. ISBN 978-2-900212-24-0.
- Silva et al. (2012). Mycologia 104(2):396-409 (doi:10.3852/11-145)
- Silva et al. (2012) Molecular Ecology 21: 2655-2670 (doi: 10.1111/j.1365-294X.2012.05557.x)
Computational Applications/Open Access Software:
- Trifusion: Software for gathering, processing and visualization of phylogenomic data.
https://github.com/OdiogoSilva/TriFusion -
ElConcatenero - Python scrip for phylogenomics analyses (doi: 10.5281/zenodo.10588)
http://zenodo.org/record/10588#.VfaiMGzYLeQ -
Phylogenomic analysis pipeline: Computation applications comprising 4 programs.